isirv Antiviral Group
Mechanisms of Resistance
Influenza A M2 inhibitors
Amantadine and rimantadine bind to a site within the pore of the M2 tetrameric channel. Single amino acid substitutions at four residues (26, 27, 30 and 31) which line the pore of the channel, and which interfere with drug binding, have been principally responsible for high level cross-resistance of natural isolates to amantadine and rimantadine. S31N is the principal substitution conferring resistance of current A(H3N2) and A(H1N1)pdm09 viruses, and of some clades of A(H5N1) viruses, although V27A, either alone or together with S31N is present in other H5N1 viruses.
Influenza Neuraminidase inhibitors (NAIs)
Reduced susceptibility of circulating influenza viruses to neuraminidase inhibitors (NAIs) is mainly due to mutations in the NA which directly reduce inhibition of enzyme activity. Mutations in the HA, which reduce receptor binding affinity and the requirement for NA, have been selected in cell culture. While some HA mutations have been seen in clinical isolates, their role in affecting drug sensitivity remains unclear.
The principal NA resistance mutations show type and subtype specificity and the level of ‘resistance’ is dependent on the inhibitor (and the assay used). The H275Y substitution close to the active site of the enzyme confers high level resistance to oseltamivir in N1 viruses (IC50 generally >1000 nM), including pre 2009 seasonal A(H1N1), A(H1N1)pdm09 and A(H5N1) viruses. Cross-resistance to peramivir is at a lower level. R292K and E119V have been responsible for high level resistance of A(H3N2) viruses to oseltamivir. Viruses with H275Y or E119V mutations retain susceptibility to zanamivir, while the R292K has a small impact on zanamivir and peramivir binding. There have been only one or two instances of selection of clinical resistance to zanamivir, due to R152K or I223R, and these have been in immunocompromised patients. Other mutations, N294S (N1 and N2) and I223V/L/R (N1) reduce susceptibility to a lesser extent, with some only affecting oseltamivir binding, while others reduce susceptibility to zanamivir and peramivir. Other mutations affecting susceptibility of influenza B isolates include: I222T, D198N, D198E, S250G and G402S.
For high affinity binding of oseltamivir and peramivir, rotation of E276 in the active site to form a salt link with R224 is necessary to form the hydrophobic pocket which accommodates the ethylpropoxy side chains of these inhibitors. The structural basis for reduced binding of oseltamivir to H275Y, N294S and R292K mutant NAs is illustrated in Figs 1, 2 and 3 below.
Although evident that high level resistance compromises clinical efficacy of the antiviral, the impact of lower level reduced susceptibility (‘resistance’) of the NA on the effectiveness of the antiviral has in general not been established.
‘Resistance’ to NI drugs is not absolute, ranging from a few fold to >1000-fold decrease in susceptibility. There are typical ranges of IC50 values that differ between influenza subtypes and between oseltamivir and zanamivir for each subtype. Furthermore, IC50 values generated by fluorescence and chemiluminescence methods should not be compared directly, as values generated by chemiluminescence assays are typically lower than those for the same virus and drug in the fluorescence test.
Influenza B viruses tend to have IC50 values for oseltamivir 10-100 fold higher than influenza A viruses. This lower susceptibility may correlate with reports of reduced clinical efficacy. For zanamivir, IC50 values for influenza B viruses are only a few-fold higher than for influenza A viruses. IC50 values for H1N1 viruses tend to be higher for zanamivir than oseltamivir, whereas the inverse is true for H3N2 viruses where IC50 values tend to be higher for oseltamivir than zanamivir.
Whilst there is no absolute definition of ‘resistance’, due, in part to lack of consensus, many define ‘resistance’ as an IC50 value at least 10 times greater than the mean IC50 for similar viruses in the same influenza season. However, given that the base line IC50 for influenza B viruses is already often 10-fold higher than for influenza A viruses, it may be more relevant to define ‘resistant’ viruses by the fold increase in IC50, relative to a base line value. This would need to be defined separately for each of the fluorescence and chemiluminescence assays.
The phylogenetic tree ![]() of NA genes of seasonal H1N1 viruses shows the relationships of oseltamivir-resistant viruses (in red), due to the H275Y substitution, which emerged during late 2007 – 2008 to contemporary oseltamivir-sensitive viruses (in black). Reference vaccine viruses are in blue. Common amino acids which distinguish the different clades are indicated (Collins et al., 2009)
of NA genes of seasonal H1N1 viruses shows the relationships of oseltamivir-resistant viruses (in red), due to the H275Y substitution, which emerged during late 2007 – 2008 to contemporary oseltamivir-sensitive viruses (in black). Reference vaccine viruses are in blue. Common amino acids which distinguish the different clades are indicated (Collins et al., 2009)
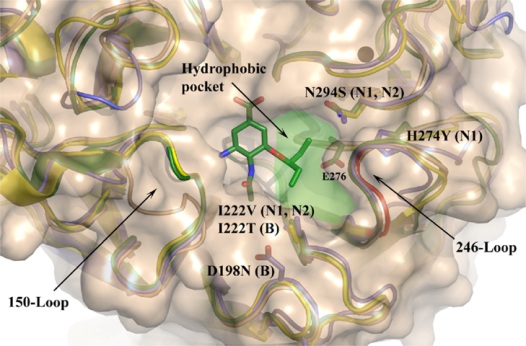
Figure 1. Locations of amino acid substitutions which reduce oseltamivir sensitivity by altering drug interaction with the hydrophobic pocket of the NA active site.
Structures of the open (gold) and closed (green) conformations of N1, and of N2 and B NAs are superimposed. A deletion of residues 244-247 in N2 is shown in red. Residues are numbered according to N2 numbering. (Hay et al., 2008).
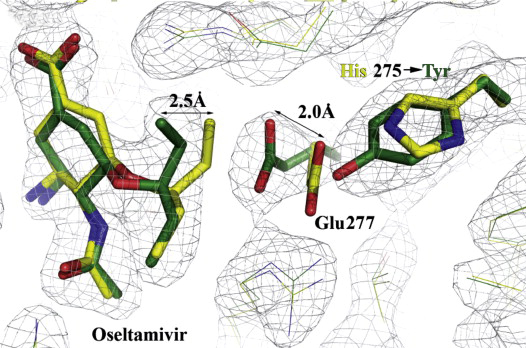
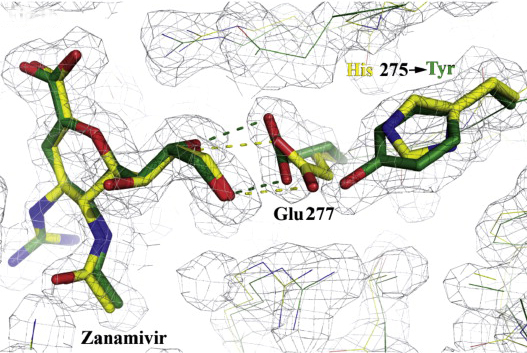
Figure 2: Structural analysis of the effects of the H275Y substitution.
X-ray structures of complexes of oseltamivir and zanamivir with wild-type (yellow) and mutant (green) N1 NAs superimposed, showing the effects of the H275Y (N1 numbering) substitution on the orientation of E277 and drug interaction (Collins et al., 2009).
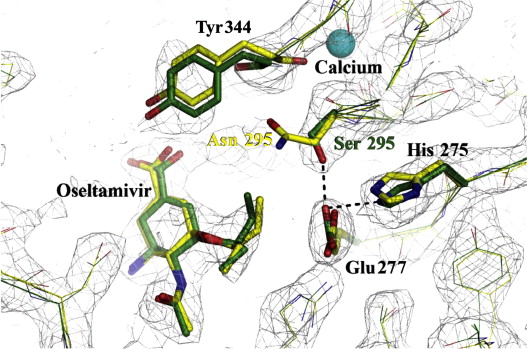
Figure 3: Structural analysis of the effects of the N295S substitution.
Superimposed X-ray structures of complexes of oseltamivir with wild-type (yellow) and mutant (green) N1 NAs showing the effect of the N295S (N1 numbering) substitution on oseltamivir interaction (Collins et al., 2009).
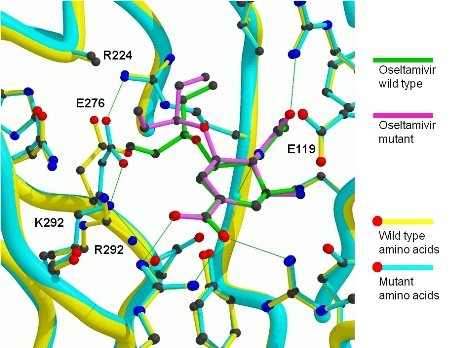
Figure 4: Structural analysis of the effects of the R292K substitution.
Superimposed images of the complexes of oseltamivir with the R292 and K292 N9 NAs, showing lack of full rotation of E276 in the K292 mutant, resulting in partial displacement of oseltamivir out of the active site. (Varghese et al., 1998)
References:
Collins, P. J., L. F. Haire, Y. P. Lin, J. Liu, R. J. Russell, P. A. Walker, S. R. Martin, R. S. Daniels, V. Gregory, J. J. Skehel, S. J. Gamblin, and A. J. Hay. 2009. Structural basis for oseltamivir resistance of influenza viruses. Vaccine 27:6317-23.
Hay, A.J., Collins, P.J. and Russell, R.J. (2008) Antivirals and Resistance. In Avian Influenza. Klenk, H-D., Matrosovich, M.N. and Steck, J., ed. Monogr Virol. 27. (Basel, Karger), pp251-270.
Varghese, J. N., P. W. Smith, S. L. Sollis, T. J. Blick, A. Sahasrabudhe, J. L. McKimm-Breschkin, and P. M. Colman. 1998. Drug design against a shifting target: a structural basis for resistance to inhibitors in a variant of influenza virus neuraminidase. Structure 6:735-46.